Model annotation¶
One of the most powerful features of CellML is its ability to import models. This means that complex models can be built up by combining previously defined models. There is a potential problem with this process, however, since the imported models (often developed by completely different modellers) may represent the same biological or biophysical entity with different expressions. The potassium channel model in A model of the potassium channel: Introducing CellML components and connections, for example, represents the intracellular concentration of potassium as ‘Ki’ (see the CellML Text code Potassium_ion_channel.cellml) but another model involving the intracellular potassium concentration may use a different expression.
The solution to this dilemma is to annotate the CellML variables with names from controlled vocabularies that have been agreed upon by the relevant scientific community. In this case we may simply want to annotate Ki as ‘the concentration of potassium in the cytosol’. This expression, however, refers to three distinct entities: concentration, potassium and cytosol. We might also want to specify that we are referring to the cytosol of a neuron … and that the neuron comes from a particular part of a giant squid (the experimental animal used by Hodgkin and Huxley). Annotations can clearly get very complicated!
What comes to our rescue here is that most scientific communities have developed controlled vocabularies together with the relationships between the terms of that vocabulary – called ontologies. Furthermore relationships can always be expressed in the form subject-predicate-object. E.g. Ki is-the-concentration-of potassium is one relationship and potassium in-the cytosol is another. Each object can become the subject of another expression. We could continue, for example, with cytosol of-the neuron, neuron of-the squid and so on. The terms s-the-concentration-of, in-the and of-the are the predicates and these semantically rich expressions too have to come from controlled vocabularies. Each of these subject-predicate-object expressions is called an RDF triple and the World Wide Web consortium [1] has established a framework called the Resource Description Framework (RDF [2]) to support these.
CellML models therefore contain two parts, one dealing with syntax (the MathML definition of the models together with the structure of components, connections, groups, units, etc) as discussed in previous sections, and one dealing with semantics (the meanings of the terms used in the models) discussed in this section [3]. This latter is also referred to as metadata – i.e. data about data.
In the CellML metadata specification [4] the first RDF subject of a triple is a CellML element (e.g. a variable such as ‘Ki’), the RDF predicate is chosen from the Biomodels Biological Qualifiers [5] list, and the RDF object is a URI (the string of characters used to identify the name of a resource [6]). Establishing these RDF links to biological and biophysical meaning is the goal of annotation.
Note the different types of subject/object used in the RDF triples: the concentration is a biophysical entity, potassium is a chemical entity, the cytosol is an anatomical entity. In fact, to cover all the terminology used in the models, CellML uses five separate ontologies:
ChEBI (Chemical Entities of Biological Interest) www.ebi.ac.uk/chebi
GO (Gene Ontology) www.geneontology.org
FMA (Foundation Model of Anatomy) fma.biostr.washington.edu/projects/fm/
Cell type ontology code.google.com/p/cell-ontology
OPB sbp.bhi.washington.edu/projects/the-ontology-of-physics-for-biology-opb
These ontologies are available through OpenCOR’s annotation facilities as explained below.
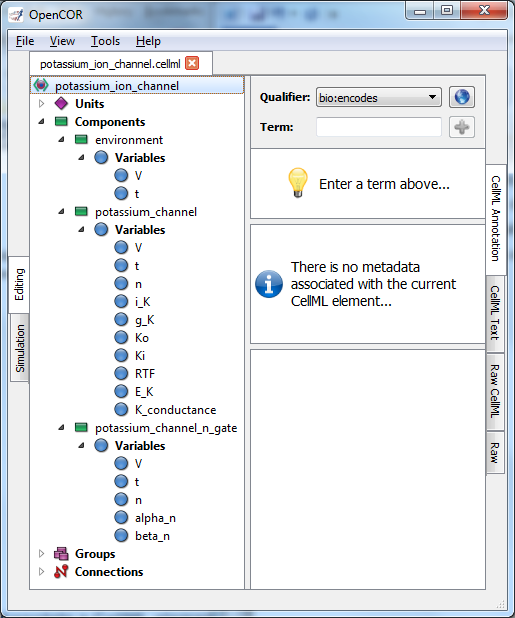
Fig. 34 Clicking on CellML Annotation lists the CellML components with their variables ready for annotation.¶
If we now go back to the potassium ion channel CellML model and, under Editing, click on CellML Annotation, the various elements of the model (Units, Components, Variables, Groups and Connections) are displayed (see Fig. 34). If you right click on any of them a popup menu will appear, which you can use to expand/collapse all the child nodes, as well as remove the metadata associated with the current CellML element or the whole CellML file. Expanding Components lists all the components and their variables. To annotate the potassium channel component, select it and specify a Qualifier from the list displayed:
bio:encodes, bio:isPropertyOf
bio:hasPart, bio:isVersionOf
bio:hasProperty, bio:occursIn
bio:hasVersion, bio:hasTaxon
bio:is, model:is
bio:isDescribedBy, model:isDerivedFrom
bio:isEncodedBy, model:isDescribedBy
bio:isHomologTo, model:isInstanceOf
bio:isPartOf, model:hasInstance
If you do not know which qualifier to use, click on the
 button to get some information about the current qualifier
(you must be connected to the internet) and go through the list of
qualifiers until you find the one that best suits your needs. Here, we
will say that you want to use bio:isVersionOf. Fig. 35 shows the
information displayed about this qualifier.
button to get some information about the current qualifier
(you must be connected to the internet) and go through the list of
qualifiers until you find the one that best suits your needs. Here, we
will say that you want to use bio:isVersionOf. Fig. 35 shows the
information displayed about this qualifier.
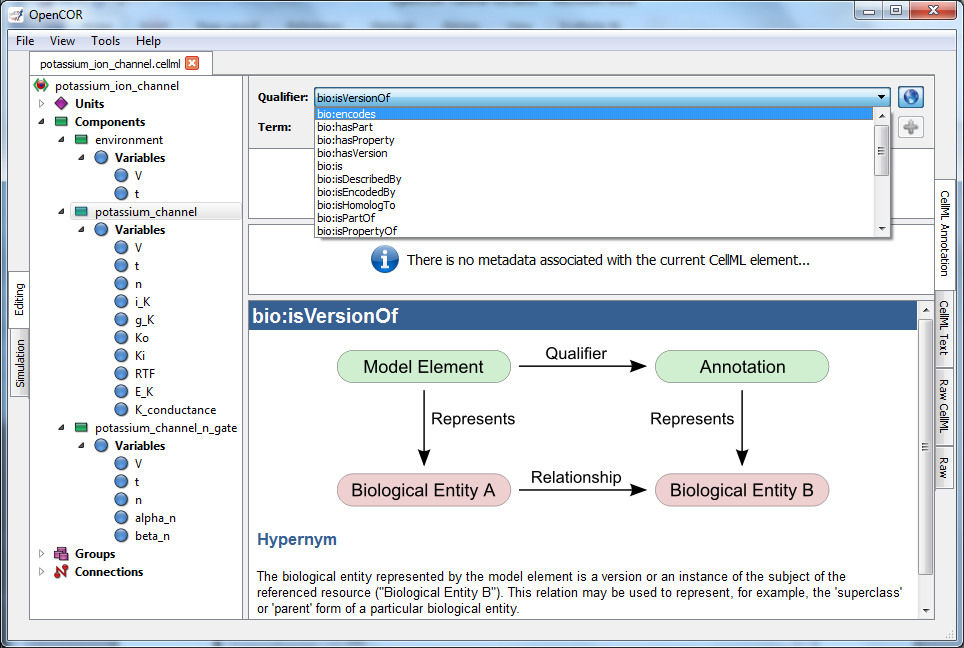
Fig. 35 The qualifiers are displayed from the top right menu. Clicking on the most appropriate one (bio:isVersionOf) gives more information about this qualifier in the bottom panel.¶
Now you need to retrieve some possible ontological terms to describe the potassium_channel component. For this you must enter a search term, which in our case is ‘potassium channel’ (note that regular expressions are supported [7]). This returns 24 possible ontological terms as shown in Fig. 36. The voltage-gated potassium channel complex is the most appropriate. Clicking on the GO identifier link shown provides more information about this term (see Fig. 37).
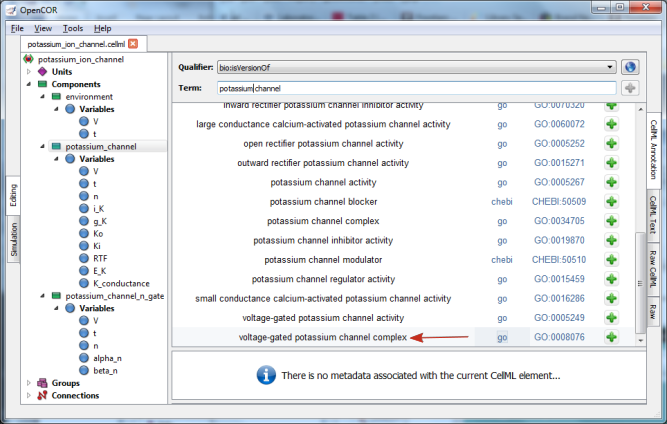
Fig. 36 The ontological terms listed when ‘potassium channel’ is entered into the search box next to Term.¶
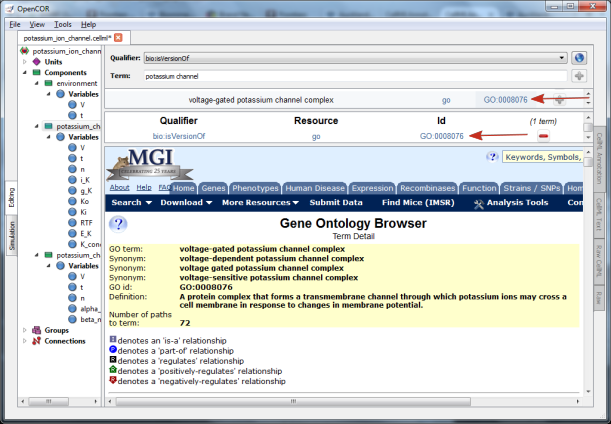
Fig. 37 The qualifier, resource & ID information in the middle
panel appears when you click on the  button next to the
selected term in Fig.32. GO identifier details are listed when either of
the arrowed links are clicked.¶
button next to the
selected term in Fig.32. GO identifier details are listed when either of
the arrowed links are clicked.¶
Now, assuming that you are happy with your choice
of ontological term, you can associate it with the potassium_channel
component by clicking on its corresponding  button which then displays
the qualifier, resource and ID information in the middle panel as shown
in Fig. 36. If you make a mistake, this can be removed by clicking on
the
button which then displays
the qualifier, resource and ID information in the middle panel as shown
in Fig. 36. If you make a mistake, this can be removed by clicking on
the  button.
button.
The first level annotation of the potassium_channel component has now been achieved. The content of the three terms in the RDF triple are shown in Fig. 38, along with the annotation for the variables Ki and Ko.
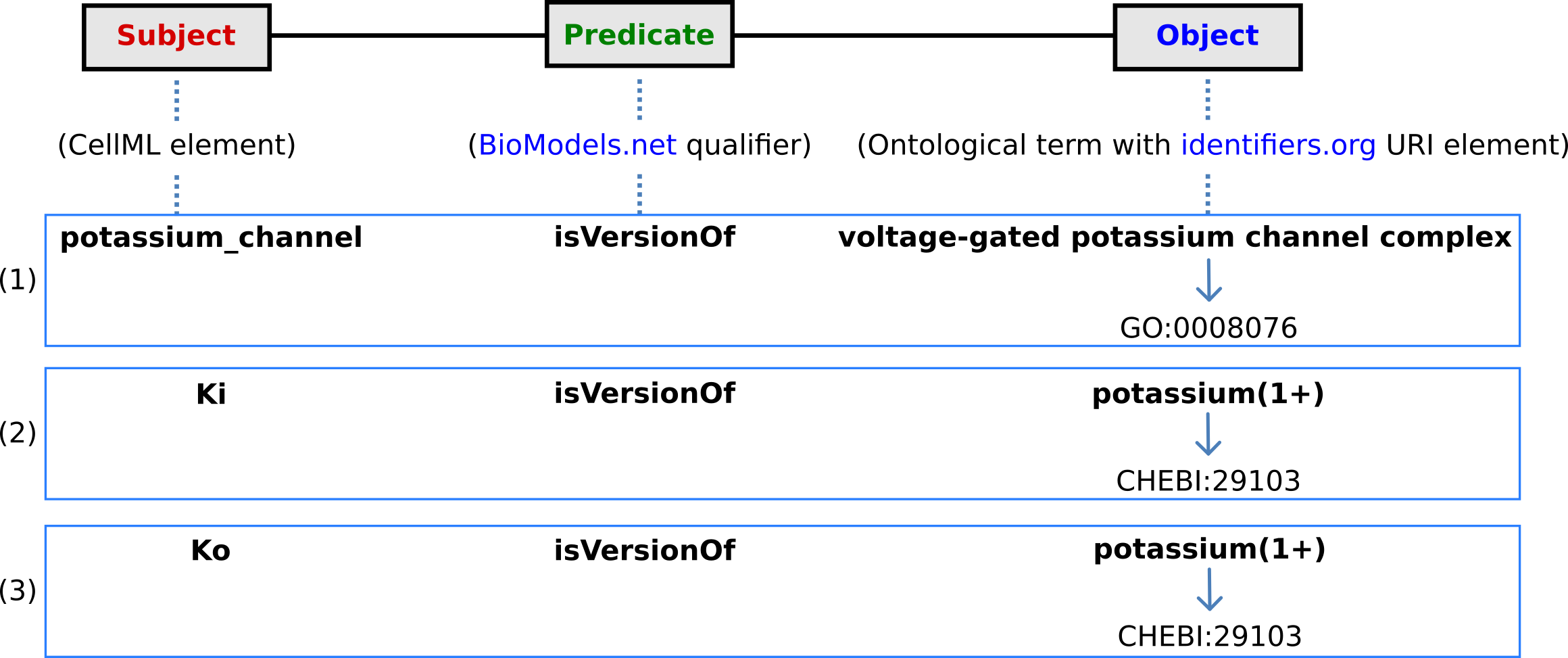
Fig. 38 The RDF triple used in CellML metadata to link a CellML element (component or variable) with an ontological term from one of the five ontologies accessed via identifiers.org, using a predicate qualifier from BioModels.net. The three examples of annotated CellML model elements shown are for (1) the potassium_channel component (this points to a GO identifier), (2) the variable Ki, and (3) the variable Ko. These two variables are defined within the potassium_channel component of the model and point to CHEBI identifiers. A further annotation is needed to identify the cellular location of those variables (since one is intracellular and one is extracellular).¶
def comp {id_000000001} potassium_channel as
var V: millivolt {pub: in, priv: out};
var t: millisec {pub: in, priv: out};
var n: dimensionless {priv: in};
var i_K: microA_per_cm2 {pub: out};
var g_K: milliS_per_cm2 {init: 36};
var {id_000000002} Ki: mM {init: 90};
var {id_000000003} Ko: mM {init: 3};
var RTF: millivolt {init: 25};
var E_K: millivolt;
var K_conductance: milliS_per_cm2 {pub: out};
E_K = RTF*ln(Ko/Ki);
K_conductance = g_K*pow(n, 4{dimensionless});
i_K = K_conductance*(V-E_K);
enddef;
When saved (the CellML Annotation tag will appear un-grayed), the result of these annotations is to add metadata to the CellML file. If you switch to the CellML Text view you will see that the elements that have been annotated appear with ID numbers, as shown above. These point to the corresponding metadata contained in the CellML file for this model and are displayed under the qualifier-resource-Id headings in the annotation window when you click on the element in the editing window.
Note that the three annotations added above are all biological annotations. Many of the other components and variables in the CellML potassium channel model deal with biophysical entities and these require the use of the OPB ontology (yet to be implemented in OpenCOR). The use of composite annotations is also being developed [8], such as “Ki is-the concentration of potassium in-the cytosol of-the neuron of-the giant-squid”, where concentration, potassium, cytosol, neuron and giant-squid are defined by the ontologies OPB, ChEBI, GO, FMA and a species ontology, respectively.
Footnotes
