The Physiome Model Repository and the link to bioinformatics¶
The Physiome Model Repository (PMR) [LCPF08] is the main online repository for the IUPS Physiome Project, providing version and access controlled repositories, called workspaces, for users to store their data. Currently there are over 700 public workspaces and many private workspaces in the repository. PMR also provides a mechanism to create persistent access to specific revisions of a workspace, termed exposures. Exposure plugins are available for specific types of data (e.g. CellML or FieldML documents) which enable customizable views of the data when browsing the repository via a web browser, or an application accessing the repository’s content via web services.
More complete documentation describing how to use PMR is available in the PMR documentation: https://models.physiomeproject.org/docs.
The CellML models on models.physiomeproject.org are listed under 20 categories, shown below: (numbers of exposures in each category are given besides the bar graph, correct as at early 2016)
Browse by category
Calcium Dynamics |
|
Cardiovascular Circulation |
|
Cell Cycle |
|
Cell Migration |
|
Circadian Rhythms |
|
Electrophysiology |
|
Endocrine |
|
Excitation-Contraction Coupling |
|
Gene Regulation |
|
Hepatology |
|
Immunology |
|
Ion transport |
|
Mechanical Constitutive Laws |
|
Metabolism |
|
Myofilament Mechanics |
|
Neurobiology |
|
pH regulation |
|
PKPD |
|
Signal Transduction |
|
Synthetic Biology |
Note that searching of models can be done anywhere on the site using the search box on the upper right hand corner. An important benefit of ensuring that the models on the PMR are annotated is that models can then be retrieved by a web-search using any of the annotated terms in the models.
To illustrate the features of PMR, click on the Hund, Rudy 2004 (Basic) model in the alphabetic listing of models under the Electrophysiology category.

Fig. 39 The Physiome Model Repository exposure page for the basic Hund-Rudy 2004 model.¶
The section labelled ‘Model Structure’ contains the journal paper abstract and often a diagram of the model[1]. This is shown for the Hund-Rudy 2004 model in Fig. 40. This model, with over 22 separate protein model components, is also a good example of why it is important to build models from modular components [CMEJ08], and in particular the individual ion channels for electrophysiology models.
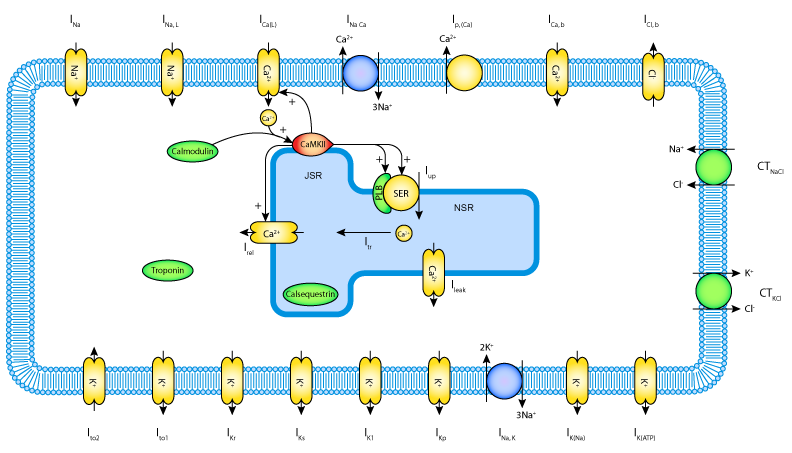
Fig. 40 A diagrammatic representation of the Hund-Rudy 2004 model.¶
There is a list of ‘Views Available’ for the CellML model on the right hand side of the exposure page. The function of each of these views is as follows:
Views Available
Documentation - Takes you to the main exposure page.
Model Metadata - Lists metadata including authors, title, journal, Pubmed ID and model annotations.
Model Curation - Provides the curation status of the model. Note: this is soon to be updated.
Mathematics - Displays all the mathematical equations contained in the model.
Generated Code - Various codes (C, C-IDA, F77, MATLAB or Python) generated from the model.
Cite this model - Provides details on how to cite use of the CellML model.
Source view - Gives a full listing of the XML code for the model.
Launch with OpenCOR - Opens the model (or simulation experiment) in OpenCOR.
Note that CellML models are available under a Creative Commons Attribution 3.0 Unported License[2]. This means that you are free to:
Share — copy and redistribute the material in any medium or format
Adapt — remix, transform, and build upon the material
for any purpose, including commercial use.
The next stage of content development for PMR is to provide a list of the modular components of these models each with their own exposure. For example, models for each of the individual ion channels used in the publication-based electrophysiological models will be available as standalone models that can then be imported as appropriate into a new composite model. Similarly for enzymes in metabolic pathways and signalling complexes in signalling pathways, etc. Some examples of these protein modules are:
Sodium/hydrogen exchanger 3 https://models.physiomeproject.org/e/236/
Thiazide-sensitive Na-Cl cotransporter https://models.physiomeproject.org/e/231/
Sodium/glucose cotransporter 1 https://models.physiomeproject.org/e/232/
Sodium/glucose cotransporter 2 https://models.physiomeproject.org/e/233/
Note that in each case, as well as the CellML-encoded mathematical model, links are provided (see Fig. 41) to the UniProt Knowledgebase for that protein, and to the Foundational Model of Anatomy (FMA) ontology (via the EMBLE-EBI Ontology Lookup Service) for information about tissue regions relevant to the expression of that protein (e.g. Proximal convoluted tubule, Apical plasma membrane; Epithelial cell of proximal tubule; Proximal straight tubule). Similar facilities are available for SMBL-encoded biochemical reaction models through the Biomodels database [AYY].
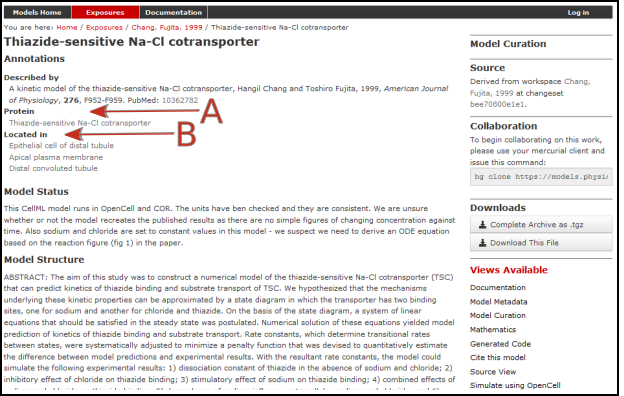
Fig. 41 The PMR workspace for the Thiazide-sensitive Na-Cl cotransporter. Bioinformatic data for this model is accessed via the links under the headings highlight by the arrows and include Protein (labelled A) and the model Location (labelled B). Other information is as already described for the Hund-Rudy 2004 model.¶
Footnotes

